
 The Philippine Coconut Authority (PCA) is planning to build a coconut oil biodiesel plant in Ivisan, Capiz (a province in the Western Visayas region) by the middle of this year in response to the growing demand for biodiesel.
News Today (Iloilo City) - April 20, 2007. The Philippine Coconut Authority (PCA) is planning to build a coconut oil biodiesel plant in Ivisan, Capiz (a province in the Western Visayas region) by the middle of this year in response to the growing demand for biodiesel.
News Today (Iloilo City) - April 20, 2007.
 Scientists working for Royal Nedalco (involved in cellulosic ethanol production), the Delft University of Technology and a firm called Bird Engineering have found a fungus in elephant dung that helped them produce a yeast strain which can efficiently ferment xylose into ethanol. The researchers consider this to be a breakthrough and see widespread application of the yeast within 5 years. More info to follow as details emerge.
Scientific American - April 19, 2007. Scientists working for Royal Nedalco (involved in cellulosic ethanol production), the Delft University of Technology and a firm called Bird Engineering have found a fungus in elephant dung that helped them produce a yeast strain which can efficiently ferment xylose into ethanol. The researchers consider this to be a breakthrough and see widespread application of the yeast within 5 years. More info to follow as details emerge.
Scientific American - April 19, 2007.
 As part of its 'Le dessous des cartes' magazine, Europe's culture TV channel ARTE airs a documentary about the geopolitics of sustainable transport tonight, at 10.20 pm CET. Readers outside of Europe can catch it here.
ARTE - April 18, 2007. As part of its 'Le dessous des cartes' magazine, Europe's culture TV channel ARTE airs a documentary about the geopolitics of sustainable transport tonight, at 10.20 pm CET. Readers outside of Europe can catch it here.
ARTE - April 18, 2007.
 Spain's diversified company the Ferry Group is investing €50 million into a biomass plantation in new EU-memberstate Bulgaria. The project will see the establishment of a 8000ha plantation of hybrid paulownia trees that will be used for the production of fuel pellets.
Dnevnik, Bulgaria - April 18, 2007. Spain's diversified company the Ferry Group is investing €50 million into a biomass plantation in new EU-memberstate Bulgaria. The project will see the establishment of a 8000ha plantation of hybrid paulownia trees that will be used for the production of fuel pellets.
Dnevnik, Bulgaria - April 18, 2007.
 Bioprocess Control signs agreement with Svensk Biogas and forms closer ties with Swedish Biogas International. Bioprocess Control develops high-tech applications that optimise the commercial production of biogas. It won Sweden's prestigious national clean-tech innovations competition MiljöInnovation 2007 for its 'Biogas Optimizer' that accelerates the biogas production process and ensures greater process stability.
NewsDesk Sweden - April 17, 2007. Bioprocess Control signs agreement with Svensk Biogas and forms closer ties with Swedish Biogas International. Bioprocess Control develops high-tech applications that optimise the commercial production of biogas. It won Sweden's prestigious national clean-tech innovations competition MiljöInnovation 2007 for its 'Biogas Optimizer' that accelerates the biogas production process and ensures greater process stability.
NewsDesk Sweden - April 17, 2007.
 A joint Bioenergy project of Purdue University and Archer Daniels Midland Company has been selected to receive funding by the U.S. Department of Energy to further the commercialization of highly-efficient yeast which converts cellulosic materials into ethanol through fermentation.
ADM - April 17, 2007. A joint Bioenergy project of Purdue University and Archer Daniels Midland Company has been selected to receive funding by the U.S. Department of Energy to further the commercialization of highly-efficient yeast which converts cellulosic materials into ethanol through fermentation.
ADM - April 17, 2007.
 Researchers at Iowa State University and the US Department of Agriculture's Agricultural Research Services (ARS) have found that glycerin, a biodiesel by-product, is as effective as conventional corn-soymeal diets for pigs.
AllAboutFeed - April 16, 2007. Researchers at Iowa State University and the US Department of Agriculture's Agricultural Research Services (ARS) have found that glycerin, a biodiesel by-product, is as effective as conventional corn-soymeal diets for pigs.
AllAboutFeed - April 16, 2007.
 U.S. demand for uranium may surge by a third amid a revival in atomic power projects, increasing concern that imports will increase and that limited supplies may push prices higher, the Nuclear Energy Institute says. Prices touched all time highs of US$113 a pound in an auction last week by a U.S producer amid plans by China and India to expand their nuclear power capacity.
International Herald Tribune - April 16, 2007. U.S. demand for uranium may surge by a third amid a revival in atomic power projects, increasing concern that imports will increase and that limited supplies may push prices higher, the Nuclear Energy Institute says. Prices touched all time highs of US$113 a pound in an auction last week by a U.S producer amid plans by China and India to expand their nuclear power capacity.
International Herald Tribune - April 16, 2007.
 Taiwan mandates a 1% biodiesel and ethanol blend for all diesel and gasoline sold in the country, to become effective next year. By 2010, the ratio will be increased to 2%.
WisconsinAg Connection - April 16, 2007. Taiwan mandates a 1% biodiesel and ethanol blend for all diesel and gasoline sold in the country, to become effective next year. By 2010, the ratio will be increased to 2%.
WisconsinAg Connection - April 16, 2007.
 Vietnam has won the prestigious EU-sponsored Energy Globe award for 2006 for a community biogas program, the Ministry of Agriculture and Rural Development announced.
ThanhNien News - April 13, 2007. Vietnam has won the prestigious EU-sponsored Energy Globe award for 2006 for a community biogas program, the Ministry of Agriculture and Rural Development announced.
ThanhNien News - April 13, 2007.
 Given unstable fossil fuel prices and their negative effects on the economy, Tanzania envisages large-scale agriculture of energy crops Deputy Minister for Agriculture, Food Security and Cooperatives, Mr Christopher Chiza has said. A 600 hectare jatropha seed production effort is underway, with the seeds expected to be distributed to farmers during the 2009/2010 growing season.
Daily News (Dar es Salaam) - April 12, 2007. Given unstable fossil fuel prices and their negative effects on the economy, Tanzania envisages large-scale agriculture of energy crops Deputy Minister for Agriculture, Food Security and Cooperatives, Mr Christopher Chiza has said. A 600 hectare jatropha seed production effort is underway, with the seeds expected to be distributed to farmers during the 2009/2010 growing season.
Daily News (Dar es Salaam) - April 12, 2007.
 Renault has announced it will launch a flex-fuel version of its Logan in Brazil in July. Brazilian autosales rose 28% to 1,834,581 in 2006 from 2004.
GreenCarCongress - April 12, 2007. Renault has announced it will launch a flex-fuel version of its Logan in Brazil in July. Brazilian autosales rose 28% to 1,834,581 in 2006 from 2004.
GreenCarCongress - April 12, 2007.
 Chevron and Weyerhouser, one of the largest forest products companies, are joining forces to research next generation biofuels. The companies will focus on developing technology that can transform wood fiber and other nonfood sources of cellulose into economical, clean-burning biofuels for cars and trucks.
PRNewswire - April 12, 2007. Chevron and Weyerhouser, one of the largest forest products companies, are joining forces to research next generation biofuels. The companies will focus on developing technology that can transform wood fiber and other nonfood sources of cellulose into economical, clean-burning biofuels for cars and trucks.
PRNewswire - April 12, 2007.
 BioConversion Blog's C. Scott Miller discusses the publication of 'The BioTown Source Book', which offers a very accessible introduction to the many different bioconversion technologies currently driving the bioenergy sector.
BioConversion Blog - April 11, 2007. BioConversion Blog's C. Scott Miller discusses the publication of 'The BioTown Source Book', which offers a very accessible introduction to the many different bioconversion technologies currently driving the bioenergy sector.
BioConversion Blog - April 11, 2007.
 China's State Forestry Administration (SFA) and the China National Cereals, Oils and Foodstuffs Import & Export Corp., Ltd. (COFCO) have signed a framework agreement over plans to cooperatively develop forest bioenergy resources, COFCO announced on its web site.
Interfax China - April 11, 2007. China's State Forestry Administration (SFA) and the China National Cereals, Oils and Foodstuffs Import & Export Corp., Ltd. (COFCO) have signed a framework agreement over plans to cooperatively develop forest bioenergy resources, COFCO announced on its web site.
Interfax China - April 11, 2007.
 The Ministry of Agriculture and Livestock of El Salvador is speeding up writing the country's biofuels law in order to take advantage of the US-Brazil cooperation agreement which identified the country as one where projects can be launched fairly quickly. The bill is expected to be presented to parliament in the coming weeks.
El Porvenir - April 11, 2007. The Ministry of Agriculture and Livestock of El Salvador is speeding up writing the country's biofuels law in order to take advantage of the US-Brazil cooperation agreement which identified the country as one where projects can be launched fairly quickly. The bill is expected to be presented to parliament in the coming weeks.
El Porvenir - April 11, 2007.
 ConocoPhillips will establish an eight-year, $22.5 million research program at Iowa State University dedicated to developing technologies that produce biofuels. The grant is part of ConocoPhillips' plan to create joint research programs with major universities to produce viable solutions to diversify America's energy sources.
Iowa State University - April 11, 2007. ConocoPhillips will establish an eight-year, $22.5 million research program at Iowa State University dedicated to developing technologies that produce biofuels. The grant is part of ConocoPhillips' plan to create joint research programs with major universities to produce viable solutions to diversify America's energy sources.
Iowa State University - April 11, 2007.
 Interstate Power and Light has decided to utilize super-critical pulverized coal boiler technology at its large (600MW) new generation facility planned for Marshalltown, Iowa. The plant is designed to co-fire biomass and has a cogeneration component. The investment tops US$1billion.
PRNewswire - April 10, 2007. Interstate Power and Light has decided to utilize super-critical pulverized coal boiler technology at its large (600MW) new generation facility planned for Marshalltown, Iowa. The plant is designed to co-fire biomass and has a cogeneration component. The investment tops US$1billion.
PRNewswire - April 10, 2007.
 One of India's largest sugar companies, the Birla group will invest 8 billion rupees (US$187 million) to expand sugar and biofuel ethanol output and produce renewable electricity from bagasse, to generate more revenue streams from its sugar business.
Reuters India - April 9, 2007. One of India's largest sugar companies, the Birla group will invest 8 billion rupees (US$187 million) to expand sugar and biofuel ethanol output and produce renewable electricity from bagasse, to generate more revenue streams from its sugar business.
Reuters India - April 9, 2007.
 An Iranian firm, Mashal Khazar Darya, is to build a cellulosic ethanol plant that will utilise switchgrass as its feedstock at a site it owns in Bosnia-Herzegovina. The investment is estimated to be worth €112/US$150 million. The plant's capacity will be 378 million liters (100 million gallons), supplied by switchgrass grown on 4400 hectares of land.
PressTv (Iran) - April 9, 2007. An Iranian firm, Mashal Khazar Darya, is to build a cellulosic ethanol plant that will utilise switchgrass as its feedstock at a site it owns in Bosnia-Herzegovina. The investment is estimated to be worth €112/US$150 million. The plant's capacity will be 378 million liters (100 million gallons), supplied by switchgrass grown on 4400 hectares of land.
PressTv (Iran) - April 9, 2007.
 The Africa Power & Electricity Congress and Exhibition, to take place from 16 - 20 April 2007, in the Sandton Convention Centre, Johannesburg, South Africa, will focus on bioenergy and biofuels.
The Statesman - April 7, 2007. The Africa Power & Electricity Congress and Exhibition, to take place from 16 - 20 April 2007, in the Sandton Convention Centre, Johannesburg, South Africa, will focus on bioenergy and biofuels.
The Statesman - April 7, 2007.
 Petrobras and Petroecuador have signed a joint performance MOU for a technical, economic and legal viability study to develop joint projects in biofuel production and distribution in Ecuador. The project includes possible joint Petroecuador and Petrobras investments, in addition to qualifying the Ecuadorian staff that is directly involved in biofuel-related activities with the exchange of professionals and technical training.
PetroBras - April 5, 2007. Petrobras and Petroecuador have signed a joint performance MOU for a technical, economic and legal viability study to develop joint projects in biofuel production and distribution in Ecuador. The project includes possible joint Petroecuador and Petrobras investments, in addition to qualifying the Ecuadorian staff that is directly involved in biofuel-related activities with the exchange of professionals and technical training.
PetroBras - April 5, 2007.
 The Société de Transport de Montréal is to buy 8 biodiesel-electric hybrid buses that will use 20% less fuel and cut 330 tons of GHG emissions per annum.
Courrier Ahuntsic - April 3, 2007. The Société de Transport de Montréal is to buy 8 biodiesel-electric hybrid buses that will use 20% less fuel and cut 330 tons of GHG emissions per annum.
Courrier Ahuntsic - April 3, 2007.
 Thailand mandates B2, a mixture of 2% biodiesel and 98% diesel. According to Energy Minister Piyasvasti Amranand, the mandate comes into effect by April next year.
Bangkok Post - April 3, 2007. Thailand mandates B2, a mixture of 2% biodiesel and 98% diesel. According to Energy Minister Piyasvasti Amranand, the mandate comes into effect by April next year.
Bangkok Post - April 3, 2007.
 In what is described as a defeat for the Bush administration, the U.S. Supreme Court ruled [*.pdf] today that environmental officials have the power to regulate greenhouse gas emissions that spur global warming. By a 5-4 vote, the nation's highest court told the U.S. Environmental Protection Agency to reconsider its refusal to regulate carbon dioxide and other emissions from new cars and trucks that contribute to climate change.
Reuters - April 2, 2007. In what is described as a defeat for the Bush administration, the U.S. Supreme Court ruled [*.pdf] today that environmental officials have the power to regulate greenhouse gas emissions that spur global warming. By a 5-4 vote, the nation's highest court told the U.S. Environmental Protection Agency to reconsider its refusal to regulate carbon dioxide and other emissions from new cars and trucks that contribute to climate change.
Reuters - April 2, 2007.
 Goldman Sachs estimates that, in the absence of current trade barriers, Latin America could supply all the ethanol required in the US and Europe at a cost of $45 per barrel – just over half the cost of US-made ethanol.
EuroToday - April 2, 2007. Goldman Sachs estimates that, in the absence of current trade barriers, Latin America could supply all the ethanol required in the US and Europe at a cost of $45 per barrel – just over half the cost of US-made ethanol.
EuroToday - April 2, 2007.
 The Kauai Island Utility Cooperative signed a long-term purchase power agreement last week with Green Energy Team, LLC. The 20-year agreement enables KIUC to purchase power from Green Energy's proposed 6.4 megawatt biomass-to-energy facility, which will use agricultural waste to generate power.
Honolulu Advertiser - April 2, 2007. The Kauai Island Utility Cooperative signed a long-term purchase power agreement last week with Green Energy Team, LLC. The 20-year agreement enables KIUC to purchase power from Green Energy's proposed 6.4 megawatt biomass-to-energy facility, which will use agricultural waste to generate power.
Honolulu Advertiser - April 2, 2007.
 The market trend to heavier, more powerful hybrids is eroding the fuel consumption advantage of hybrid technology, according to a study done by researchers at the University of British Columbia.
GreenCarCongress - March 30, 2007. The market trend to heavier, more powerful hybrids is eroding the fuel consumption advantage of hybrid technology, according to a study done by researchers at the University of British Columbia.
GreenCarCongress - March 30, 2007.
 Hungarian privately-owned bio-ethanol project firm Mabio is planning to complete an €80-85 million ethanol plant in Southeast Hungary's Csabacsud by end-2008.
Onet/Interfax - March 29, 2007. Hungarian privately-owned bio-ethanol project firm Mabio is planning to complete an €80-85 million ethanol plant in Southeast Hungary's Csabacsud by end-2008.
Onet/Interfax - March 29, 2007.
 Energy and engineering group Abengoa announces it has applied for planning permission to build a bioethanol plant in north-east England with a capacity of about 400,000 tonnes a year.
Reuters - March 29, 2007. Energy and engineering group Abengoa announces it has applied for planning permission to build a bioethanol plant in north-east England with a capacity of about 400,000 tonnes a year.
Reuters - March 29, 2007.
 The second European Summer School on Renewable Motor Fuels will be held in Warsaw, Poland, from 29 to 31 August 2007. The goal of the event is to disseminate the knowledge generated within the EU-funded RENEW (Renewable Fuels for Advanced Powertrains) project and present it to the European academic audience and stakeholders. Topics on the agenda include generation of synthetic gas from biomass and gas cleaning; transport fuel synthesis from synthetic gas; biofuel use in different motors; biomass potentials, supply and logistics, and technology, cost and life-cycle assessment of BtL pathways.
Cordis News - March 27, 2007. The second European Summer School on Renewable Motor Fuels will be held in Warsaw, Poland, from 29 to 31 August 2007. The goal of the event is to disseminate the knowledge generated within the EU-funded RENEW (Renewable Fuels for Advanced Powertrains) project and present it to the European academic audience and stakeholders. Topics on the agenda include generation of synthetic gas from biomass and gas cleaning; transport fuel synthesis from synthetic gas; biofuel use in different motors; biomass potentials, supply and logistics, and technology, cost and life-cycle assessment of BtL pathways.
Cordis News - March 27, 2007.
 Green Swedes want even more renewables, according to a study from Gothenburg University. Support for hydroelectricity and biofuels has increased, whereas three-quarters of people want Sweden to concentrate more on wind and solar too. Swedes still back the nuclear phase-out plans. The country is Europe's largest ethanol user. It imports 75% of the biofuel from Brazil.
Sveriges Radio International - March 27, 2007. Green Swedes want even more renewables, according to a study from Gothenburg University. Support for hydroelectricity and biofuels has increased, whereas three-quarters of people want Sweden to concentrate more on wind and solar too. Swedes still back the nuclear phase-out plans. The country is Europe's largest ethanol user. It imports 75% of the biofuel from Brazil.
Sveriges Radio International - March 27, 2007.
 Fiat will launch its Brazilian-built flex-fuel Uno in South Africa later this year. The flex-fuel Uno, which can run on gasoline, ethanol or any combination of the two fuels, was displayed at the Durban Auto Show, and is set to become popular as South Africa enters the ethanol era.
Automotive World - March 27, 2007. Fiat will launch its Brazilian-built flex-fuel Uno in South Africa later this year. The flex-fuel Uno, which can run on gasoline, ethanol or any combination of the two fuels, was displayed at the Durban Auto Show, and is set to become popular as South Africa enters the ethanol era.
Automotive World - March 27, 2007.
 Siemens Power Generation (PG) is to supply two steam turbine gensets to a biomass-fired plant in Três Lagoas, 600 kilometers northwest of São Paulo. The order, valued at €22 million, was placed by the Brazilian company Pöyry Empreendimentos, part of VCP (Votorantim Celulose e Papel), one of the biggest cellulose producers in the Americas.
PRDomain - March 25, 2007. Siemens Power Generation (PG) is to supply two steam turbine gensets to a biomass-fired plant in Três Lagoas, 600 kilometers northwest of São Paulo. The order, valued at €22 million, was placed by the Brazilian company Pöyry Empreendimentos, part of VCP (Votorantim Celulose e Papel), one of the biggest cellulose producers in the Americas.
PRDomain - March 25, 2007.
 Asia’s demand for oil will nearly double over the next 25 years and will account for 85% of the increased demand in 2007, Organization of Petroleum Exporting Countries (Opec) officials forecast yesterday at a Bangkok-hosted energy conference.
Daily Times - March 24, 2007. Asia’s demand for oil will nearly double over the next 25 years and will account for 85% of the increased demand in 2007, Organization of Petroleum Exporting Countries (Opec) officials forecast yesterday at a Bangkok-hosted energy conference.
Daily Times - March 24, 2007.
 Portugal's government expects total investment in biomass energy will reach €500 million in 2012, when its target of 250MW capacity is reached. By that date, biomass will reduce 700,000 tonnes of carbon emissions. By 2010, biomass will represent 5% of the country's energy production.
Forbes - March 22, 2007. Portugal's government expects total investment in biomass energy will reach €500 million in 2012, when its target of 250MW capacity is reached. By that date, biomass will reduce 700,000 tonnes of carbon emissions. By 2010, biomass will represent 5% of the country's energy production.
Forbes - March 22, 2007.
 The Scottish Executive has announced a biomass action plan for Scotland, through which dozens of green energy projects across the region are set to benefit from an additional £3 million of funding. The plan includes greater use of the forestry and agriculture sectors, together with grant support to encourage greater use of biomass products.
Energy Business Review Online - March 21, 2007. The Scottish Executive has announced a biomass action plan for Scotland, through which dozens of green energy projects across the region are set to benefit from an additional £3 million of funding. The plan includes greater use of the forestry and agriculture sectors, together with grant support to encourage greater use of biomass products.
Energy Business Review Online - March 21, 2007.

|
|
|
|
|
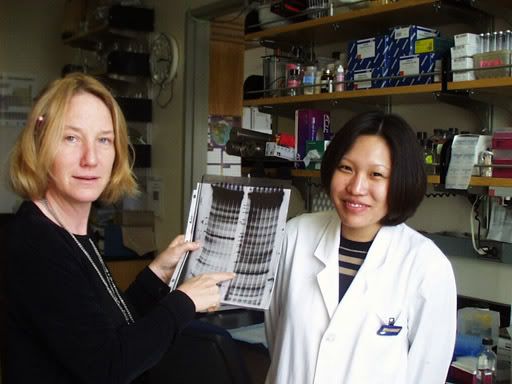 The bacterium Syntrophus aciditrophicus, one of the most extreme-survival organisms ever discovered, has had its genome sequenced. Microbiologists think the findings on the extremophile's unique metabolism may be used in the production of biohydrogen.
The bacterium Syntrophus aciditrophicus, one of the most extreme-survival organisms ever discovered, has had its genome sequenced. Microbiologists think the findings on the extremophile's unique metabolism may be used in the production of biohydrogen. bioenergy :: biofuels :: energy :: sustainability :: bacterium :: microbe :: genome :: biohydrogen :: biomass ::
bioenergy :: biofuels :: energy :: sustainability :: bacterium :: microbe :: genome :: biohydrogen :: biomass ::  --------------
--------------
 The Philippine Coconut Authority (PCA) is planning to build a coconut oil biodiesel plant in Ivisan, Capiz (a province in the Western Visayas region) by the middle of this year in response to the growing demand for biodiesel.
The Philippine Coconut Authority (PCA) is planning to build a coconut oil biodiesel plant in Ivisan, Capiz (a province in the Western Visayas region) by the middle of this year in response to the growing demand for biodiesel.








0 Comments:
Post a Comment
Links to this post:
Create a Link
<< Home